Creating the Examplomycin Dataset
This vignette demonstrates how to create a simulated pharmacokinetic
dataset for a fictional drug called examplomycin. The dataset is
designed to showcase the capabilities of the admr package
for aggregate data modeling.
Overview
The examplomycin dataset is a simulated pharmacokinetic study with the following characteristics: - 500 healthy subjects - Single oral dose of 100 mg - 9 sampling time points per subject - Two-compartment model with first-order absorption - Random effects on all parameters - Proportional residual error
Data Generation Function
The generate_data function creates the examplomycin
dataset with the following features:
Two-compartment model with first-order absorption
Log-normal random effects on all parameters
Proportional residual error
Standardized sampling schedule
nlmixr2-compatible data format
Show code
generate_data <- function(n, times, seed = 1) {
set.seed(seed)
# Define the pharmacokinetic model
mod <- function(){
model({
# Parameters
ke = cl / v1 # Elimination rate constant
k12 = q / v1 # Rate constant for central to peripheral transfer
k21 = q / v2 # Rate constant for peripheral to central transfer
# Differential equations for drug amount in compartments
d/dt(depot) = -ka * depot
d/dt(central) = ka * depot - ke * central -
k12 * central +
k21 * peripheral
d/dt(peripheral) = k12 * central -
k21 * peripheral
# Concentration in the central compartment
cp = central / v1
})
}
mod <- rxode2(mod)
mod <- mod$simulationModel
# Population parameters
theta <- c(cl = 5, v1 = 10, v2 = 30, q = 10, ka = 1)
# Correlation matrix for random effects
omegaCor <- matrix(c(1, 0, 0, 0, 0,
0, 1, 0, 0, 0,
0, 0, 1, 0, 0,
0, 0, 0, 1, 0,
0, 0, 0, 0, 1),
dimnames = list(NULL, c("eta.cl", "eta.v1", "eta.v2", "eta.q",
"eta.ka")), nrow = 5)
# Standard deviations of random effects
iiv.sd <- c(0.3, 0.3, 0.3, 0.3, 0.3)
# Create covariance matrix
iiv <- iiv.sd %*% t(iiv.sd)
omega <- iiv * omegaCor
# Generate random effects
mv <- mvrnorm(n, rep(0, dim(omega)[1]), omega)
# Create individual parameters
params.all <-
data.table(
"ID" = seq(1:n),
"cl" = theta['cl'] * exp(mv[, 1]),
"v1" = theta['v1'] * exp(mv[, 2]),
"v2" = theta['v2'] * exp(mv[, 3]),
"q" = theta['q'] * exp(mv[, 4]),
"ka" = theta['ka'] * exp(mv[, 5])
)
# Create event table
ev <- et() %>%
et(amt = 100) %>% # Single dose
et(0) %>% # Initial time point
et(times) %>% # Sampling schedule
et(ID = seq(1, n)) %>% # Subject IDs
as.data.frame()
# Solve the model
sim <- rxSolve(mod, events = ev, iCov = params.all, cores = 0, addCov = T) %>%
mutate(ID = as.integer(id), TIME = as.numeric(time)) %>%
dplyr::select(-c(id, time)) %>%
mutate(AMT = ifelse(TIME == 0, 100, 0)) %>%
mutate(EVID = ifelse(TIME == 0, 101, 0)) %>%
mutate(CMT = ifelse(TIME == 0, 1, 2))
# Add residual error
sim$rv <- rnorm(nrow(sim), 0, 0.2)
sim$DV <- round(sim$cp * (1 + sim$rv), 3)
sim <- merge(sim, params.all)
# Select final columns
dat <- sim %>%
dplyr::select("ID", "TIME", "DV", "AMT", "EVID", "CMT")
return(dat)
}Generating the Dataset
We’ll generate the examplomycin dataset with: - 500 subjects - 9 time points (0.1, 0.25, 0.5, 1, 2, 3, 5, 8, 12 hours) - Random seed for reproducibility
examplomycin <- generate_data(
n = 500, # Number of subjects
times = c(.1, .25, .5, 1, 2, 3, 5, 8, 12), # Sampling times
seed = 1 # Random seed
)## ℹ parameter labels from comments are typically ignored in non-interactive mode## ℹ Need to run with the source intact to parse comments
head(examplomycin)## ID TIME DV AMT EVID CMT
## 1 460 0.00 0.000 100 101 1
## 2 460 0.10 0.752 0 0 2
## 3 460 0.25 1.932 0 0 2
## 4 460 0.50 3.694 0 0 2
## 5 460 1.00 3.479 0 0 2
## 6 460 2.00 4.003 0 0 2Saving the Dataset
The dataset is saved as a package data object for use in other vignettes and examples:
Visualizing the Dataset
We’ll create a concentration-time plot to visualize the simulated data:
# Create concentration-time plot
ggplot(examplomycin, aes(x = TIME, y = DV, color = factor(ID))) +
geom_line(alpha = 0.7) + # Connect points with lines
geom_point(size = 2, alpha = 0.8) + # Add observation points
scale_color_viridis_d(name = "Subject ID") + # Color by subject
labs(
title = "Concentration-Time Profile",
x = "Time (hours)",
y = "Observed Concentration (DV)"
) +
theme_minimal() +
theme(legend.position = "none") # Hide legend for clarity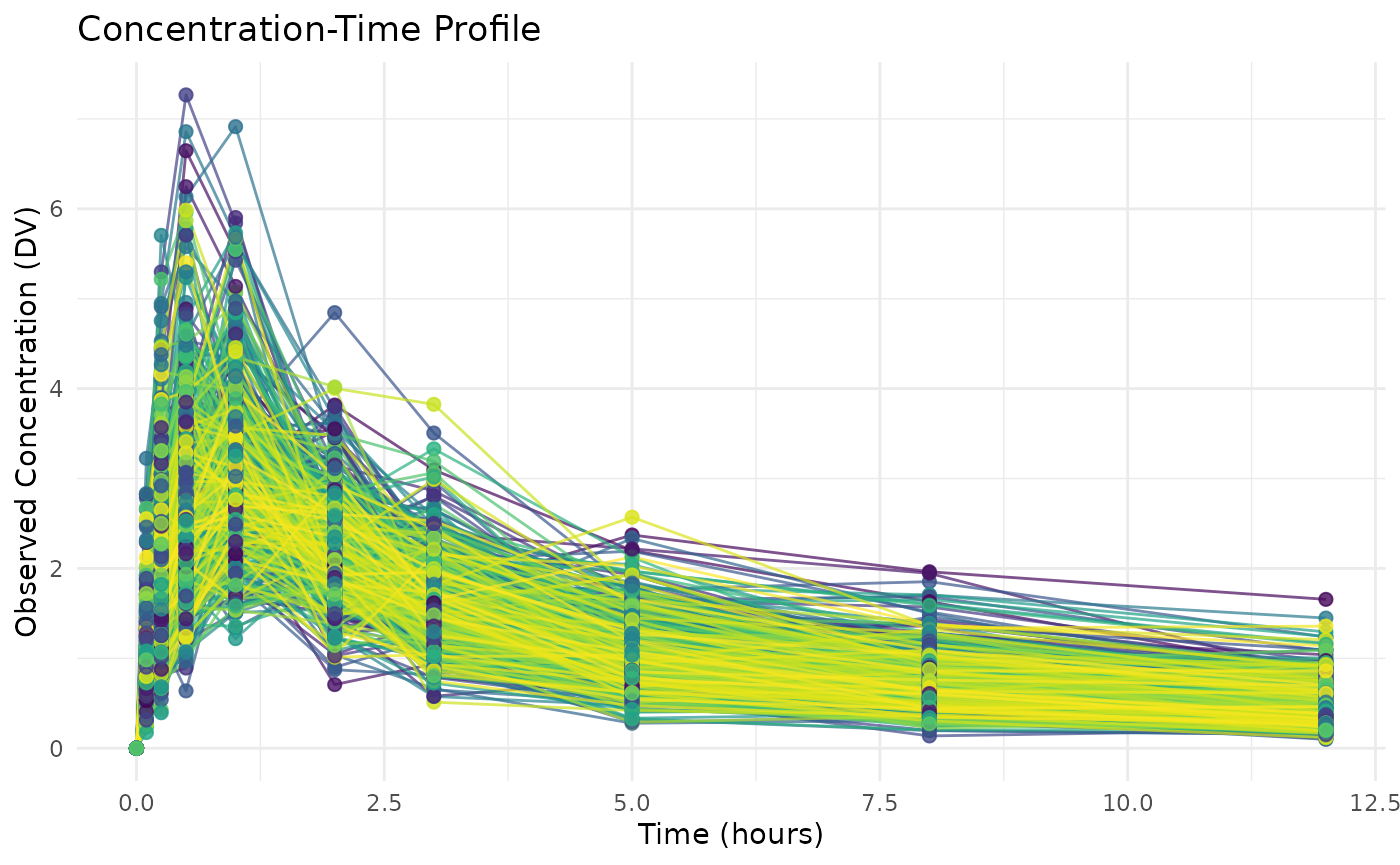
Dataset Structure
The examplomycin dataset contains the following columns:
ID: Subject identifierTIME: Observation time (hours)DV: Observed concentrationAMT: Dose amount (100 mg or NA)EVID: Event type (101 for dose, 0 for observation)CMT: Compartment number (1 for depot, 2 for central)
This dataset serves as a realistic example for demonstrating
aggregate data modeling techniques in the admr package.
